In the Laboratory of Therapeutic Nucleic Acids, we perform the synthesis of modified nucleosides, nucleotides and oligonucleotides which are the models for advanced physico-chemical, structural and biological studies. Modified RNA oligonucleotides, mimicking fragments of tRNA containing natural modified nucleosides, are suitable models for the structure-function relationship analysis. DNA models modified with boron clusters provide a platform for the synthesis of functional nanostructures with potential use in the treatment of cancer and civilization diseases. We are looking for active therapeutic nucleic acids exhibiting the properties of inhibitors of gene expression (antisense oligonucleotides, siRNA) or inhibitors of protein enzyme activity (aptamers). We conduct research on the synthesis and function of nucleotide analogs (nucleoside triphosphates, dinucleoside-polyphosphates) involved in cell metabolic processes. We are also interested in the function of proteins interacting with nucleic acids, involved in biosynthesis of modified nucleosides found in tRNA, and proteins from the HIT family interacting with nucleotide signal molecules.
Area of interest:
The nucleoside modifications, present at the wobble position of anticodon sequence of transfer ribonucleic acid (tRNA), have been the objects of our interest for many years. Nucleosides in this position play an important role in proper reading of genetic information in the process of protein biosynthesis. In particular, we are interested in 5-substituted 2-thiouridines (R5S2U), found only in three types of tRNA: specific for lysine, glutamic acid or glutamine (tRNALys, tRNAGlu and tRNAGln). In studies carried out together with the Prof. Elzbieta Sochacka team (TUL), we have found that in the oxidative environment at in vitro conditions, the 2-thiouridine is desulfured, i.e. the sulfur atom is removed from the molecule, and the products of this reaction are uridine (U), naturally occurring in RNA and a deprived of sulfur atom nucleoside called 4-pyrimidinone riboside (H2U), the hydrogen bond acceptors and donors pattern of which is different than that of uridine and 2-thiouridine. The presence of the H2U modification can be considered a damage since tRNAs containing the H2U unit may do not exert its function properly. Currently, we aim to clarify whether process of 2-thiouridine desulfuration also occurs under natural conditions, in the eukaryotic cells subjected to oxidative stress.
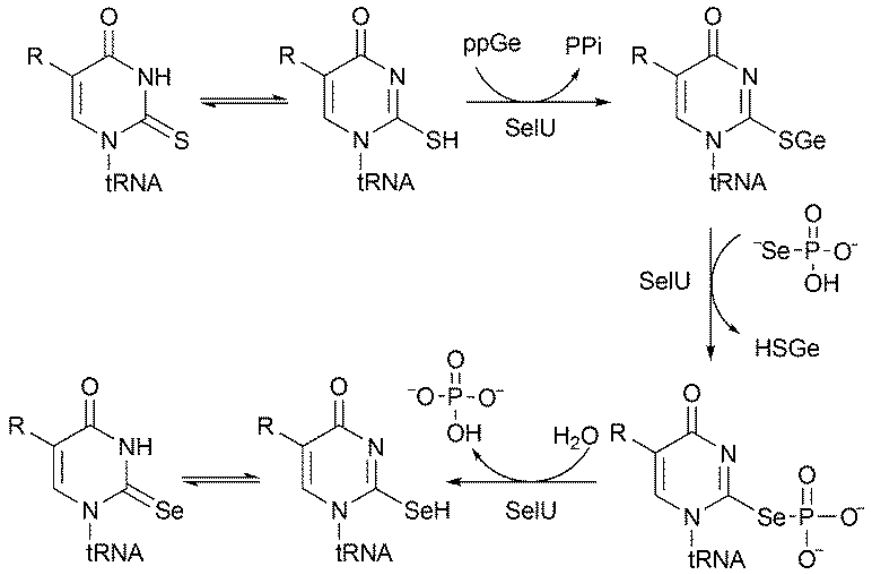
Recently, our group discovered the mechanism of transformation of 2-thiouridine (S2U) to 2-selenouridine (Se2U) present in the RNA chain. We proved that in the first step S2U is converted into S-geranyl-2-thiouridine, which is an intermediate in the synthesis of Se2U (Scheme below). Both of these processes are catalyzed by the bacterial enzyme tRNA 2-selenoridine synthase (SelU). We are currently conducting research aimed at explaining at the molecular level the function of selenium in nucleosides occurring in the wobble position of tRNA.
The mechanism of transformation of 2-thiouridune-tRNA to 2-selenouridine-tRNA, discovered in our studies was recently published in the FEBS Letters journal at 2018 (Sierant et al. FEBS Lett. 592 (13), 2248-2258, 2018).
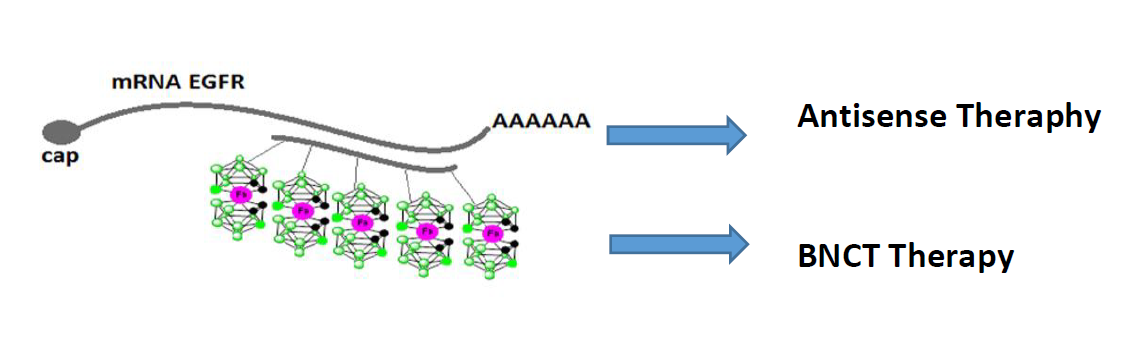
In collaboration with Institute of Medical Biology of PAS, Lodz (Prof. dr hab. Z. Lesnikowski’s team) we are conducting research on synthesis of a new type of bioorganic composites consisting of nucleic acids (DNA) and boron clusters, their physico-chemical properties and their potential as building blocks to create programmable nanostructures.
As a part of the research we synthesized modified antisense oligonucleotides (ASO) directed towards the EGF receptor gene, post-synthetically conjugated with carboranyl residues and spectrally characterized by HPLC, MS, UV, CD. The implementation of the project requires also synthesis of “oligopodal” building blocks (DNA-oligopods / oligofunctionalized boron clusters) using solid-phase method, study of their physicochemical and biochemical characteristics, as well as testing their ability to create 2D / 3D nanostructures and topology of this particals (AFM, Cryo-TEM). The biological studies of the obtained compounds and nanostructures are also important part of the project (including assessment of the exo- and endogenous level of EGFR protein in cancer cells lines, assessment of cytotoxicity, proliferative changes, nucleolytic stability and oxidative stress).
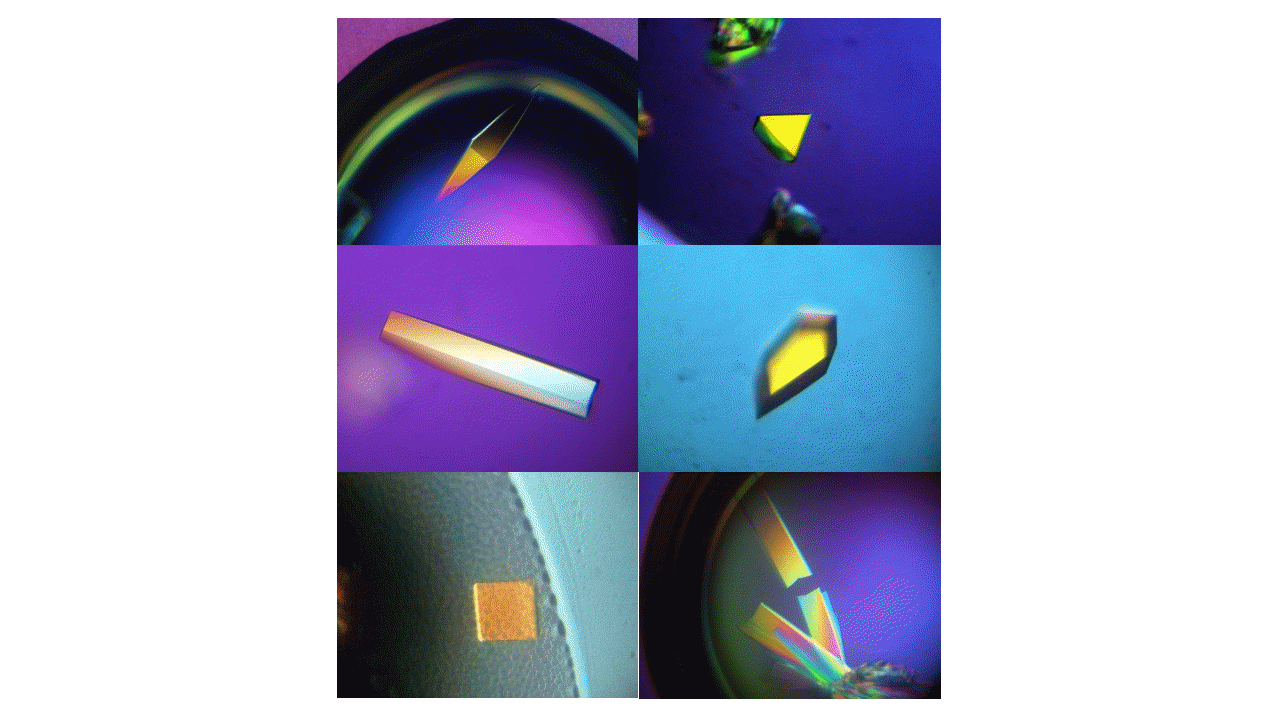
The research covers crystallization, single crystal diffraction experiments, and solving and refinement of the obtained structures of nucleic acids, oligonucleotides, proteins (mainly from the HIT proteins family) and their complexes with selected ligands (e.g. a modified thrombin binding aptamer complexed with thrombin, HINT1 protein with Ap4A non-hydrolysable analog) and low molecular compounds, e.g. nucleoside and nucleotide derivatives and other biologically active compounds, e.g. potential drugs. Understanding a complete structure of the studied molecules and the manner of intermolecular interactions allows, for example, to determine the mechanism of action of drugs or mechanisms of enzymatic activity of selected proteins. In our Institute, it is possible to conduct diffraction experiments using synchrotron radiation on DESY (Hamburg, Germany) and BESSY (Berlin, Germany) synchrotrons. Currently, CMMS is preparing for the opening of a X-ray analysis laboratory equipped with X-ray diffractometers for single crystal, as well as for and a powder diffraction measurements.
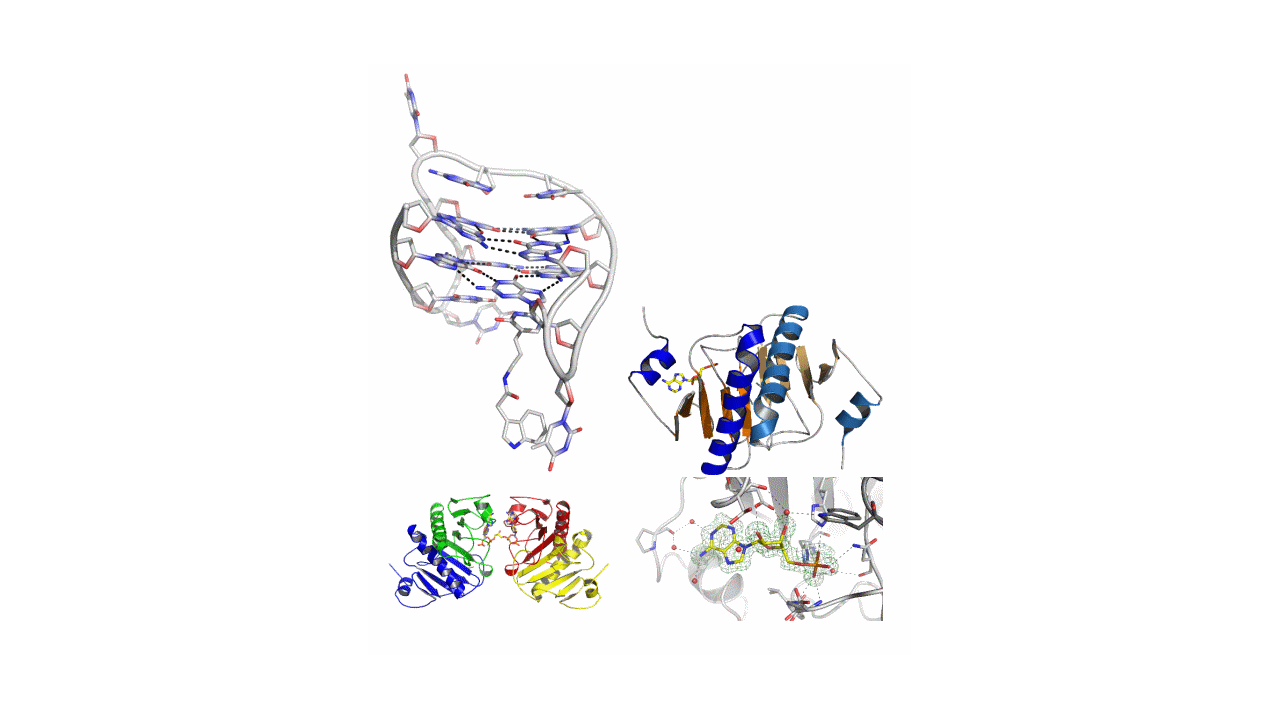
At this topic, the mechanism of apoptosis induced by the Fhit protein was elucidated using non-hydrolyzable Ap4A analogs (originally designed and synthesized in house) and which are inhibitors of the enzymatic activity of the studied protein (Krakowiak et al., Bio. Med. Chem. 11 (2011) 5053). In addition, a new region of interaction between the Fhit protein and the substrate/substrate analog has been found (Krakowiak et al. FEBS Lett. 591 (2017), 548).
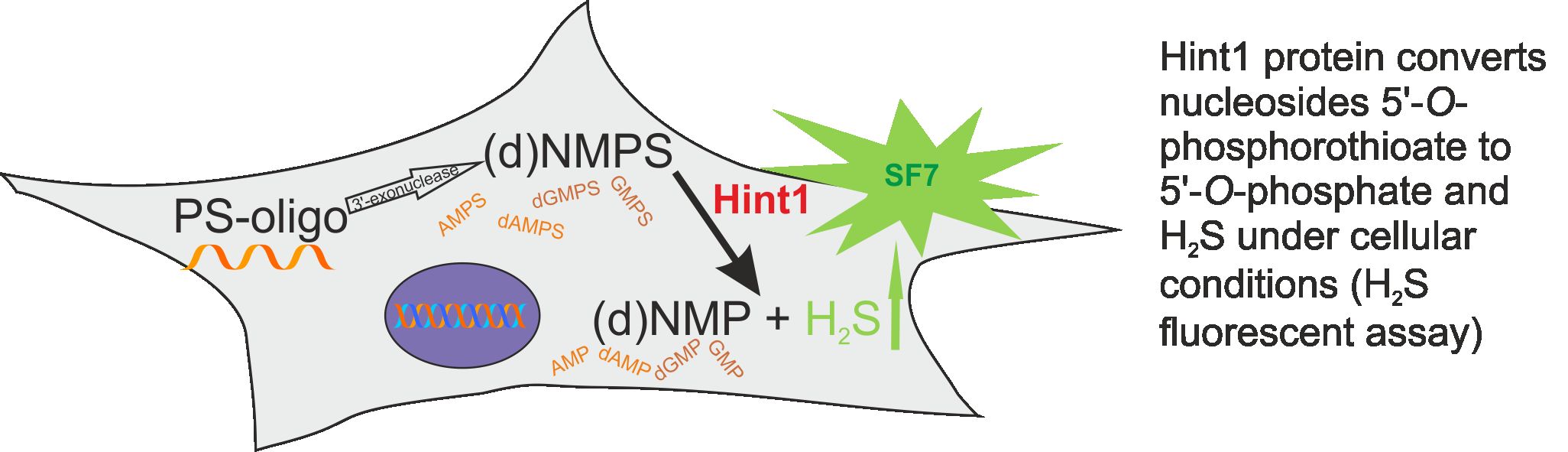
In our earlier studies on Hint1 phosphoramidase we determined the stereochemistry of the P-N bond hydrolysis in nucleoside derivatives of tiophosphoramidate (Krakowiak et al. Chem.Comm. 2007, 2163). Moreover, Hint1 was also found as desulfurase of nucleoside 5′-O-thiophosphates and in this reaction the release of hydrogen sulfide was observed (Ozga et al. J. Biol. Chem. 285 (2010), 40809). Connection of H2S with etiology of many diseases under physiological conditions (e.g. hypertension, Alzheimer’s disease) is known. Further studies have shown that the above desulfuration reaction also occurs inside the cells with participation of the Hint1 enzyme (Figure below, Krakowiak et al. BBA, 1840 (2014) 3357; Krakowiak et al., Biochem. Phar. 163 (2019) 250). This can be important both, in the antisense therapy with phosphorothioate analogs of oligonucleotides, as well as in the possibility of application of phosphorothioate nucleoside analogs as H2S donors in the treatment of some diseases.